A way to identify neurons by shape
Introduce
This algorithm is utlized by identifying neurons, specially when the traditional nuclear counting method can't work. For example, sometime neurons' nuclear is hard to stain but the satellite are stained domiantlyby DIPI. Moreover, the amount of images are limited so Deep Learning may not work well by the insufficient images. Therefore, this algorithm tries to identify nuerons by shape, with a fewer samples.
process:
The algorithm are consistued by three main steps:
(1)analyzing the labeled samples, all samples are come from a images with axis paris. meanwhile, calculating pixels distribution function.
(2)Scan images in the "Image" folder. In a image, find all points where the pixels meet pixels distribution function. Then selected its near by area to calculate the binary image.
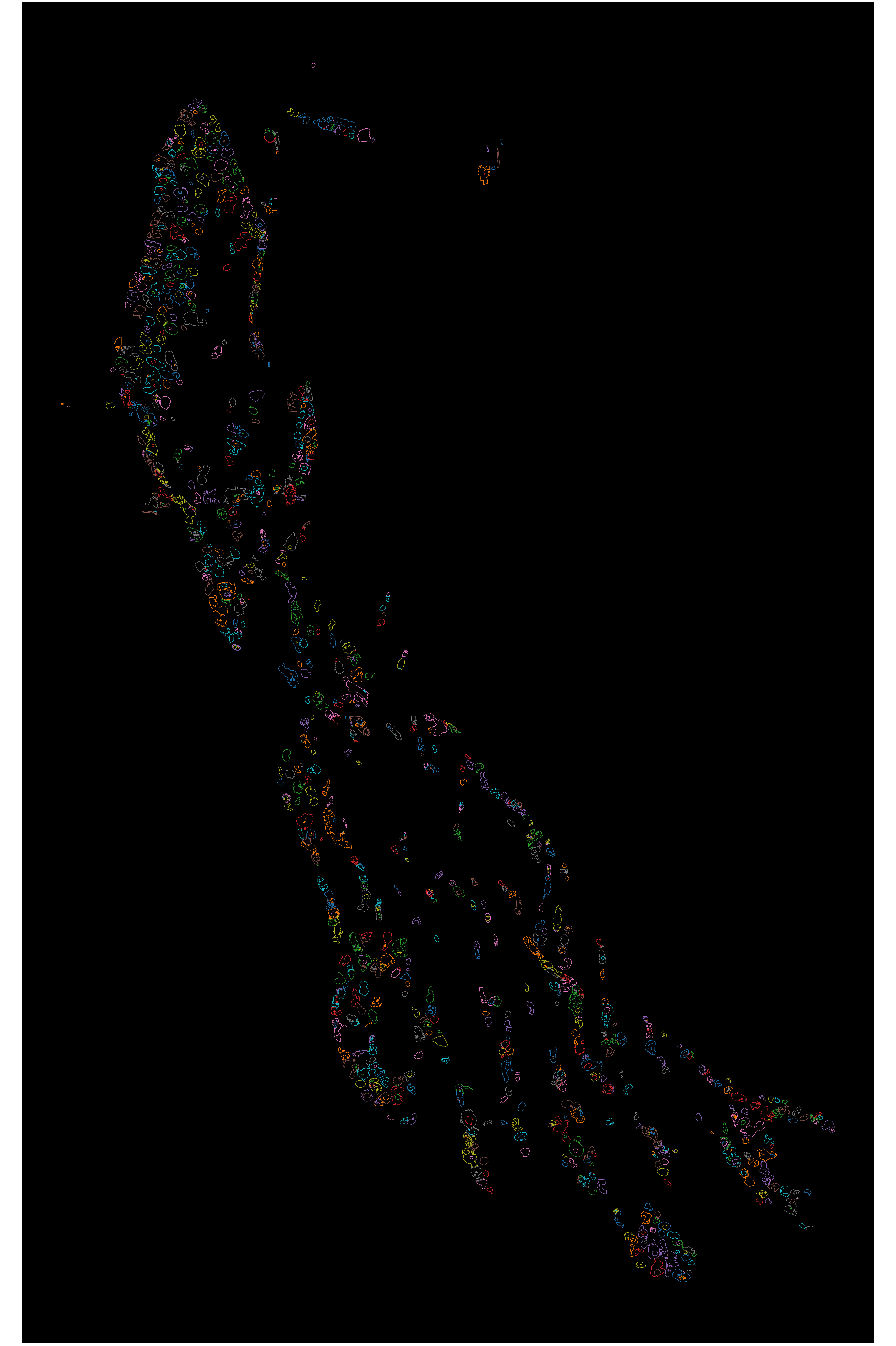
(3)combine all section to as one image, then compare the all the shape of connected component with the labels', eventually get the quantity of neurons.
How to use
1.put label image to the "labeled_image" folder, and input the coordinate of the selecting labels in the "large_high_pixel_labels" file.
2.put images which need to analaze in the "image" folder
3.run the shell start.py.
Parameters
.......................................................................
omit | parameter | explain
-s | --section_scale | the size of section (0.1 ~ 10)
-l | --lower_threshold | lower bound of threshold basing on label (0.1 ~ 10)
-u | --upper_threshold | lower bound of threshold basing on label (0.1 ~ 10)
-e | --erosion | the size of erosion mask (1 ~ 10)
-m | --median | the size of median mask (1 ~ 10)
.......................................................................
example:
python start.py -s 2.0 -l 0.6Result