Haogao Gu, Leo L.M. Poon
This work was conducted in School of Public Health, The University of Hong Kong under the supervison of Prof. Leo Poon.
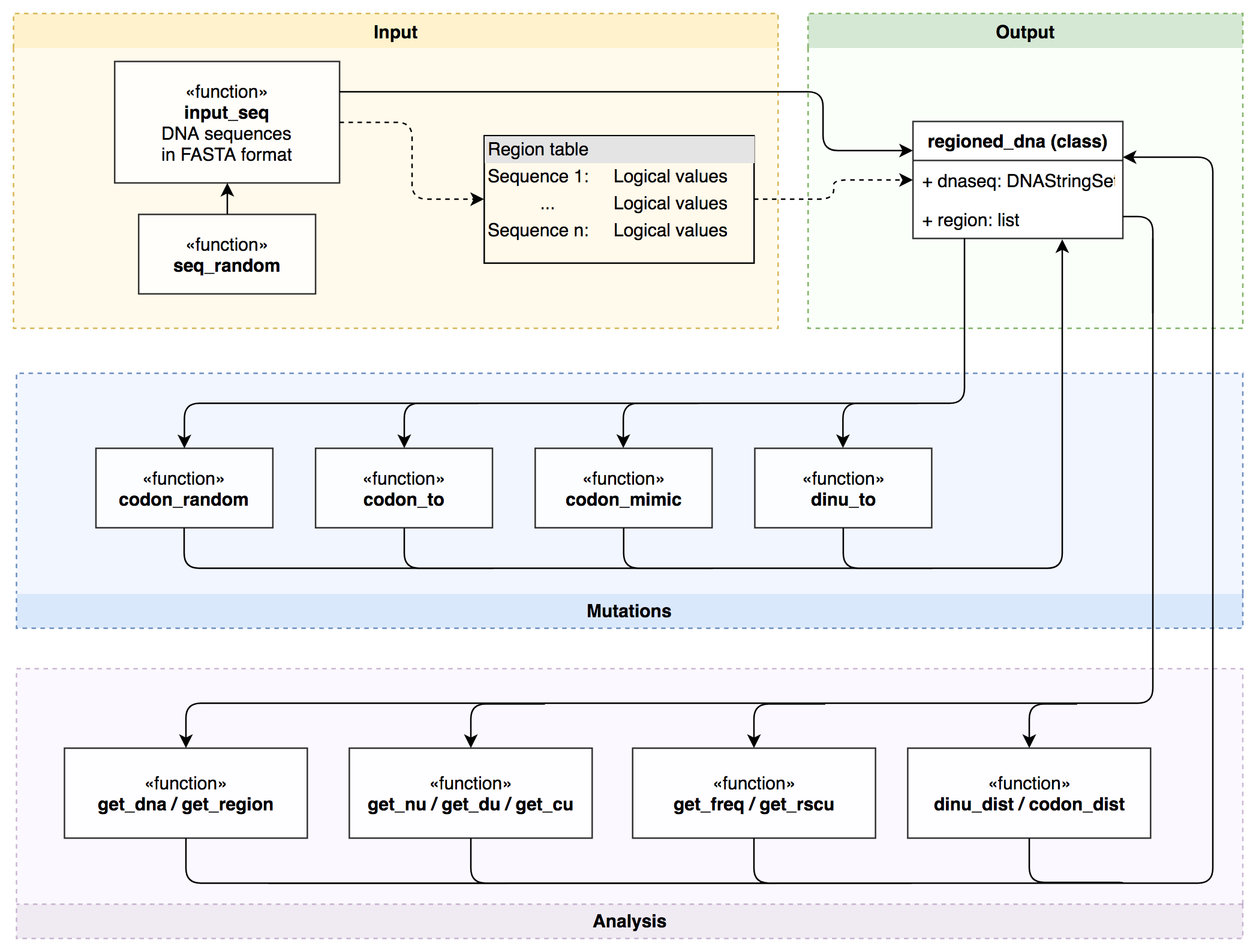
SynMut designs synonymous mutants for DNA sequences.
There are increasing demands on designing virus mutants with specific dinucleotide or codon composition. This tool can take both dinucleotide preference and/or codon usage bias into account while designing mutants. It also works well for desinging mutants with extremely over-/under- represented dinucleotides.
This tool was originally designed for generating recombinant virus sequences in influenza A virus to study the effects of different dinucleotide usage and codon usage, yet the functions provided in this package can be generic to a variety of other biological researches.
Use the below code to install the package.
# Stable version
if (!requireNamespace("BiocManager"))
install.packages("BiocManager")
if (!requireNamespace("SynMut"))
BiocManager::install("SynMut")
# Development version
if (!requireNamespace("devtools"))
install.packages("devtools")
if (!requireNamespace("SynMut"))
devtools::install_github("Koohoko/SynMut")Details tutorial please refer to the vignette.
The strategies and functionalities of the codom_mimic and dinu_to functions can be found at here.
https://bioconductor.org/packages/devel/bioc/html/SynMut.html
Changes in version 1.1.5 (2022-06-03)
- Enhancement: add other non-standard genetic codes for functions codon_random, dinu_to, codon_mimic.
Changes in version 1.1.4 (2020-11-12)
- bug fix: fix for function codon_mimic.
Changes in version 1.1.3 (2019-05-08)
- Revise dinu_to.keep algorithm, enhance performance.
Changes in version 1.1.2 (2019-05-08)
- Bug fix: "dinu_to" ifelse issue in get_optimal_codon.
Changes in version 1.1.1 (2019-05-06)
- Bug fix: "dinu_to" fix wrong result with "keep == TRUE" parameter.