conda create -n sunet2 python=3.8
pip install -U scikit-learn
pip3 install torch torchvision torchaudio --index-url https://download.pytorch.org/whl/cu118python predict.py -hemi left -l 7 -i /mnt/d/Spherical_U-Net/examples/left_hemisphere/40962/test1.lh.40k.vtk -o 0715.vtkThis is the code for paper "Spherical U-Net on Cortical Surfaces: Methods and Applications".
- Linux
NVIDIA GPUCUDA CuDNN
- python (3.8)
- torch (2.2.1)
- torchvision (0.17.1)
- pyvista (0.43.4)
- numpy (1.21.5)
- scikit-learn (1.3.2)
- vtk (9.0.3)
- python (3.6)
- pytorch (0.4.1+)
- torchvision (0.2.1+)
- tensorboardx (1.6+)
- pyvista (0.22.4+)
You can use conda to easily create an environment for the experiment using following command:
conda create -n sunet python=3.6
conda activate sunet
conda install pytorch torchvision cudatoolkit=10.0 -c pytorch
conda install -c conda-forge pyvista
Some basic tools can be found in the Spherical U‐Net package and installed from PyPI:
pip install sphericalunet
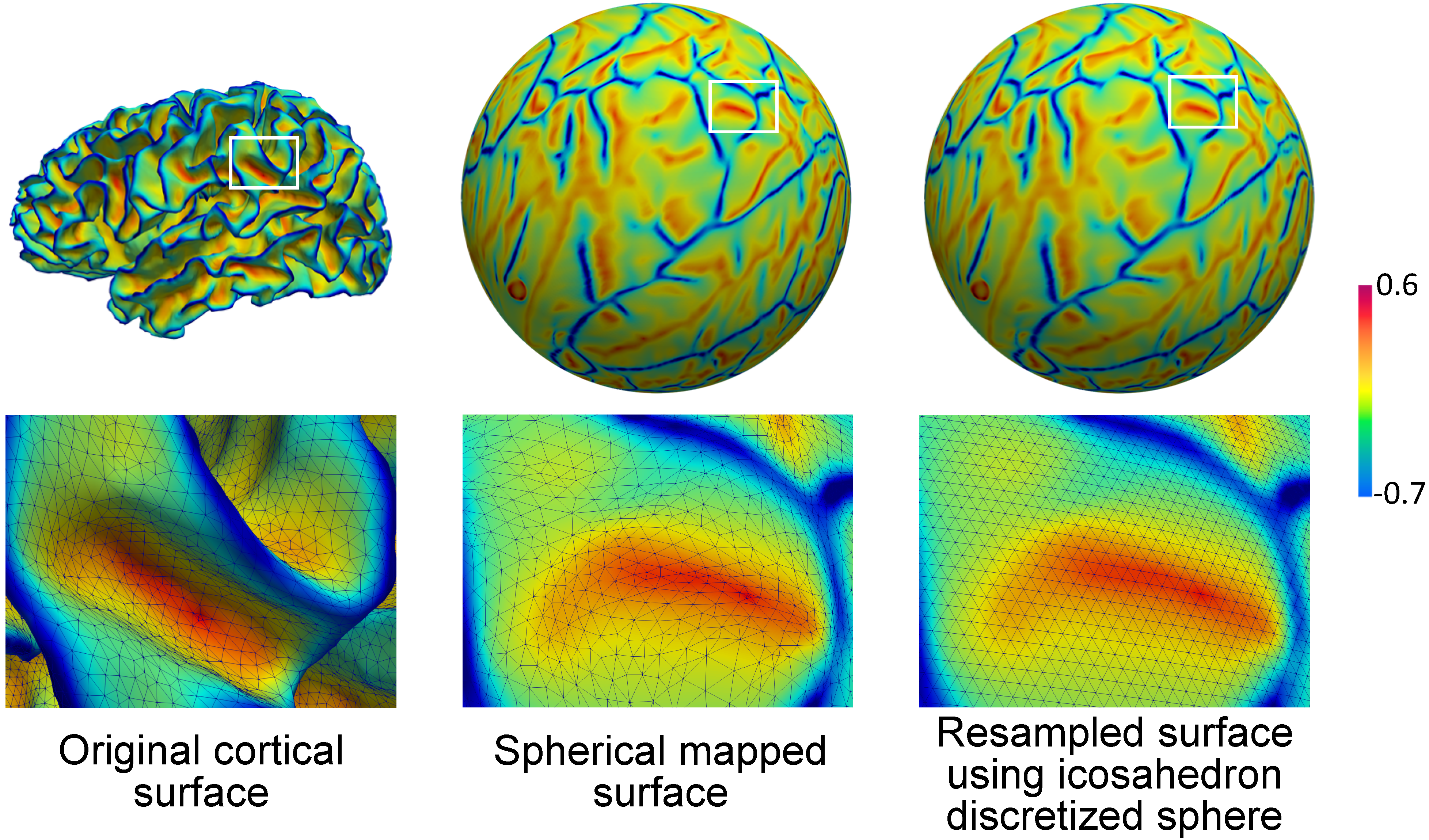
The input file is a cortical inner surface of one hemisphere in vtk format reconstructed from neuroimaging pipelines [1][2], which has been resampled as either 40,962 or 163,842 vertices. Two features, i.e., mean curvature and average convexity, are required for the parcellation, denoted as “curv” and “sulc” field attributes in the vtk file, respectively. For resampling and feature computing, FreeSurfer [2] can be used. To be consistent with the trained model, gyral crests should have negative curvature values, while sulcal bottoms should have positive curvature values.
After data prepration, modify the train.py file to match the training data in your own path. Then, run:
python train.py
You can easily obtain the output parcellation maps on your surfaces via the following commands. To predict a single surface’ parcellation map:
python predict.py -hemi left -l 7 -i input.vtk -o output.vtk
To predict the parcellation maps of multiple surfaces in the same folder:
python predict.py -hemi left -l 7 -in_folder in_folder -out_folder out_folder
You can also view the help information of the whole usage of this command by running
python predict.py -h
Usage: predict.py [-h] [--hemisphere {left,right}] [--level {7,8}]
[--input INPUT] [--in_folder INPUT_FOLDER] [--output OUTPUT] [--out_folder OUT_FOLDER]
Predict the parcellation maps with 36 regions from the input surfaces
optional arguments:
-h, --help show this help message and exit
--hemisphere {left,right}, -hemi {left,right}
Specify the hemisphere for parcellation, left or
right. (default: left)
--level {7,8}, -l {7,8}
Specify the level of the surfaces. Generally, level
7 spherical surface is with 40962 vertices, 8 is with
163842 vertices. (default: 7)
--input INPUT, -i INPUT
filename of input surface (default: None)
--in_folder INPUT_FOLDER, -in_folder INPUT_FOLDER
folder path for input files. Will parcelalte all the
files end in .vtk in this folder. Accept input or
in_folder. (default: None)
--output OUTPUT, -o OUTPUT
Filename of ouput surface. (default: [input].parc.vtk)
--out_folder OUT_FOLDER, -out_folder OUT_FOLDER
folder path for ouput surface. Accept output or
out_folder. (default: [in_folder])
Troubleshoot notes:
- The code requires
inputorin_folderoption, not both, for single surface’ parcellation or all surfaces in the folder. - The input data should be end in .vtk.
You can test the code using the example surfaces we provided in the examples folder. Simply run:
python predict.py -hemi left -l 7 -i examples/left_hemisphere/40962/test1.lh.40k.vtk
You will get the corresponding output surface at the same folder with name test1.lh.40k.parc.vtk.
Or, run the command for all the 3 surface in the same folder:
python predict.py -hemi left -l 7 -in_folder examples/left_hemisphere/40962
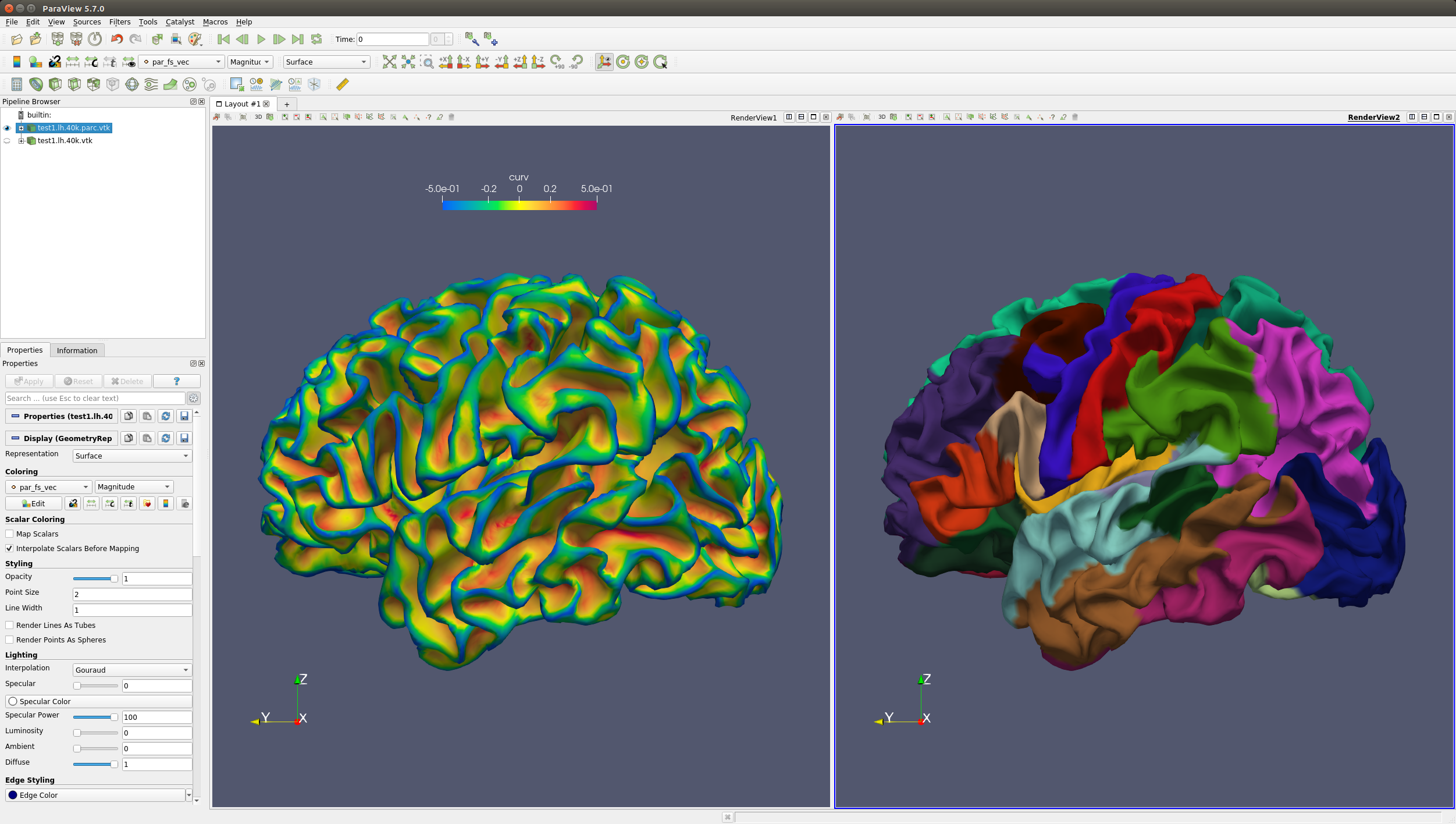
You can use Paraview software to visualize the parcellated surface in VTK format. An example of the input curvature map and output parcellation map are shown below. More usages about Paraview please refer to Paraview.
 .
.
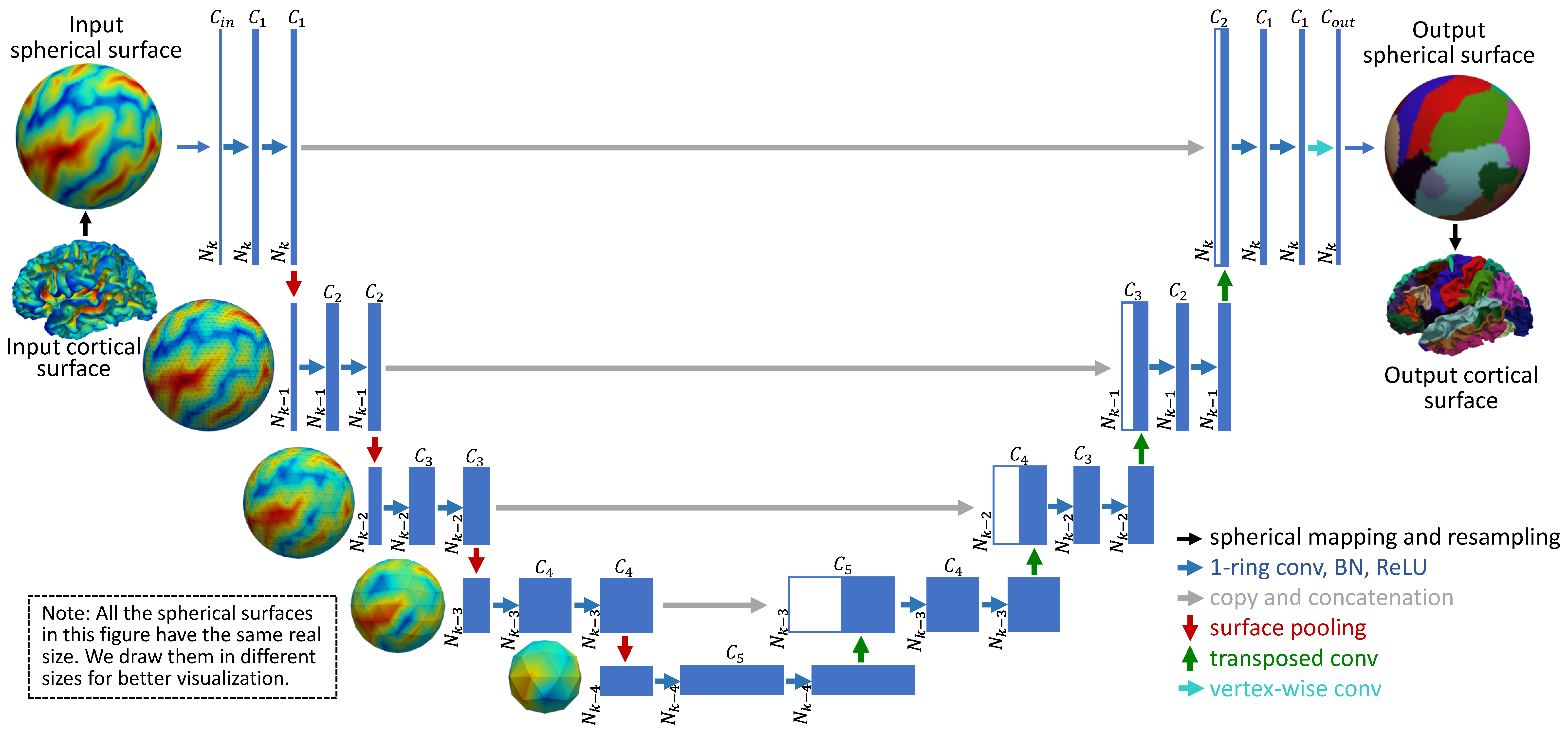
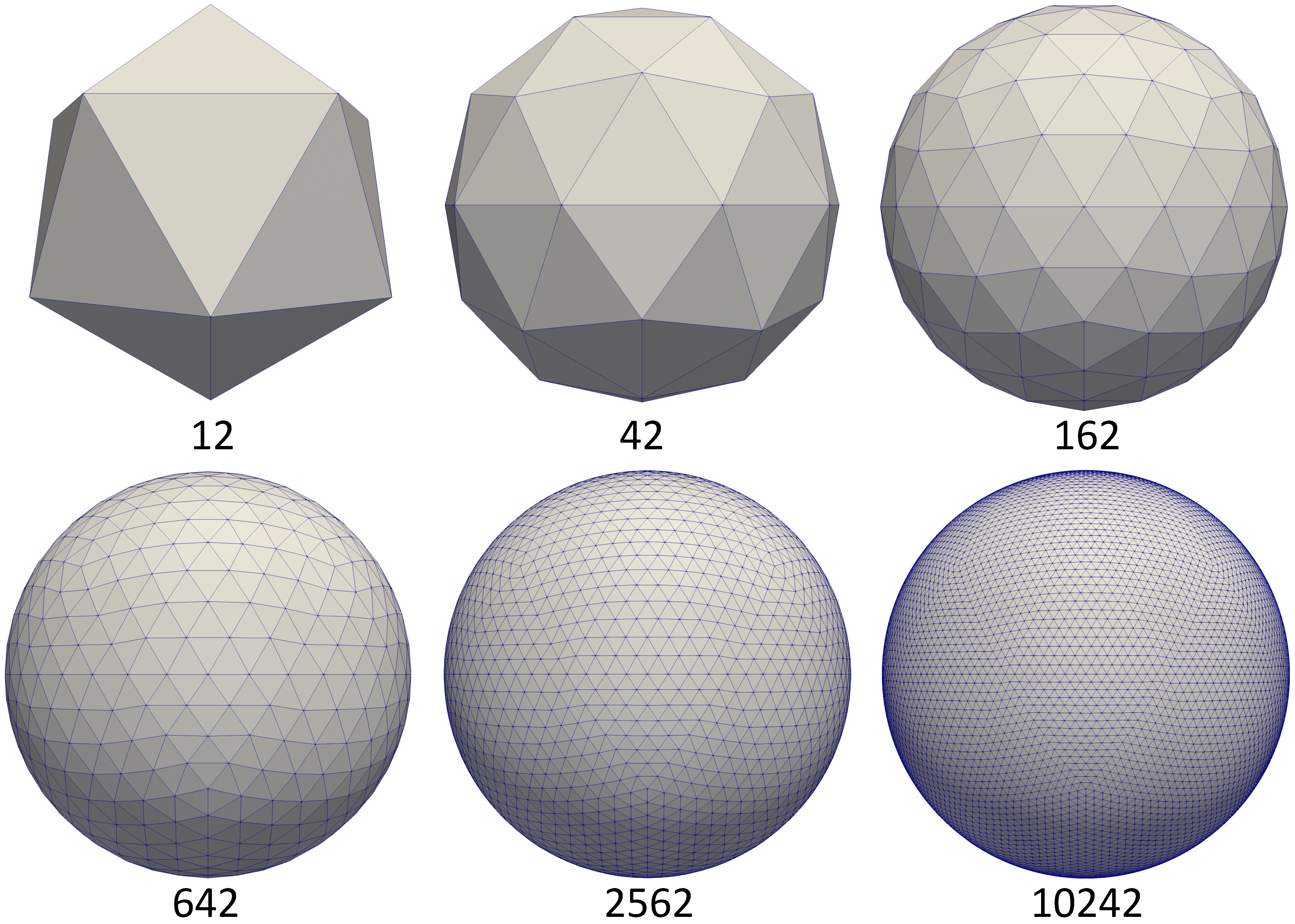
The brain cortical surface lie in a non-Euclidean space represented by triangular meshes.
The convolution on the spherical surface is performed baed on the 1-ring filter
 .
.
We provide 3 types of filter on the spherical surfaces.
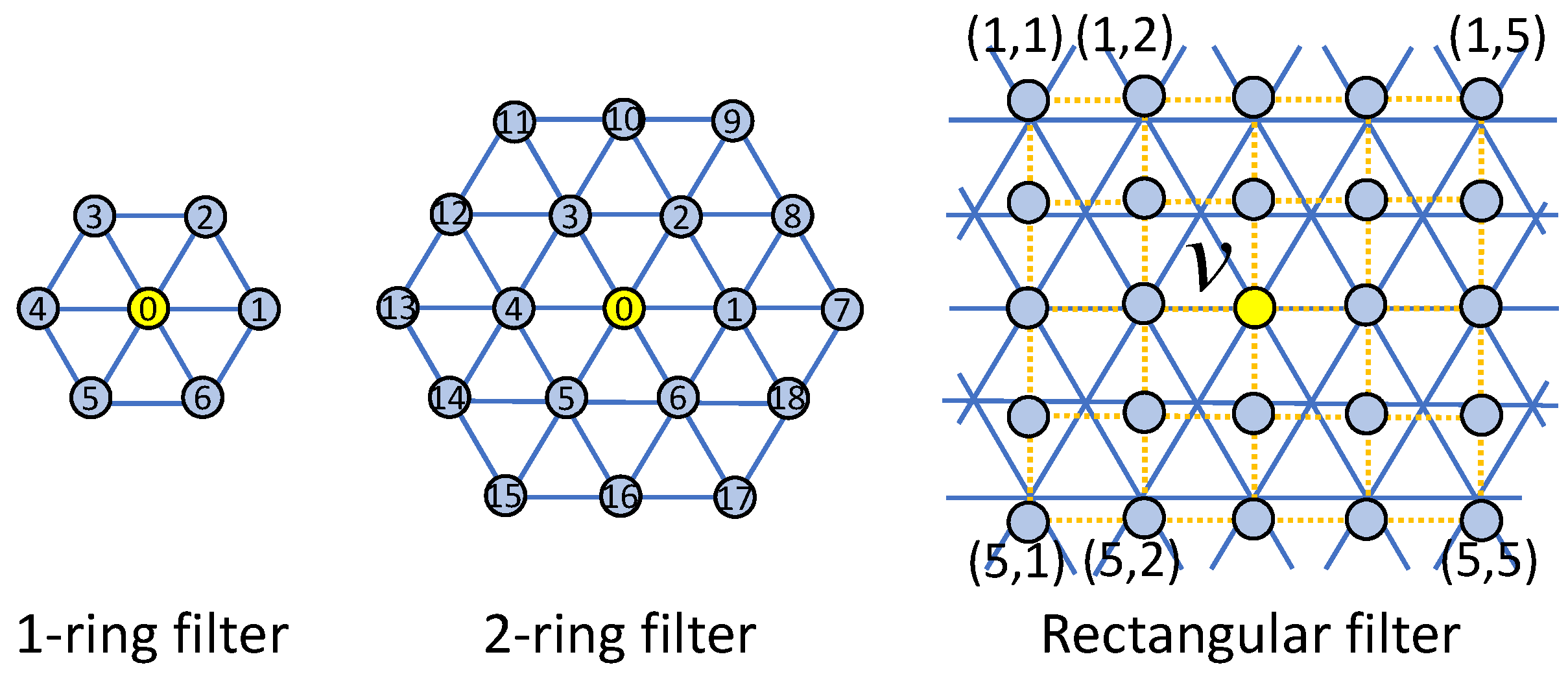 .
.
If you use this code for your research, please cite as:
Fenqiang Zhao, et.al. Spherical U-Net on Cortical Surfaces: Methods and Applications. Information Processing in Medical Imaging (IPMI), 2019.
Fenqiang Zhao, et.al. Spherical Deformable U-Net: Application to Cortical Surface Parcellation and Development Prediction. IEEE Transactions on Medical Imaging, 2021.
-
check_sdf.py calculate SDF value on the sphere and then add to surf vtk
-
mv_hemi_npy.py archive npz files to six folders, lh & rh
/home/cxc0366/fsl/lib/python3.11/site-packages/torch/nn/modules/loss.py:101: UserWarning: Using a target size (torch.Size([40962])) that is different to the input size (torch.Size([40962, 1])). This will likely lead to incorrect results due to broadcasting. Please ensure they have the same size. return F.l1_loss(input, target, reduction=self.reduction)
torch.cuda.OutOfMemoryError: CUDA out of memory. Tried to allocate 87.51 GiB (GPU 0; 47.50 GiB total capacity; 29.21 MiB already allocated; 46.95 GiB free; 46.00 MiB reserved in total by PyTorch) If reserved memory is >> allocated memory try setting max_split_size_mb to avoid fragmentation. See documentation for Memory Management and PYTORCH_CUDA_ALLOC_CONF
Problem found in file: ./Test4/lh/fold1/130922.lh.InnerSurf.RegByFS.Resp40962_linemask_skeleton.npz Problem found in file: ./Test4/lh/fold5/130114.lh.InnerSurf.RegByFS.Resp40962_linemask_skeleton.npz